Loading required package: ggplot2Loading required package: lattice
Attaching package: 'kernlab'The following object is masked from 'package:ggplot2':
alphainTrain <- createDataPartition(y=spam$type,
p=0.75, list=FALSE)
training <- spam[inTrain,]
testing <- spam[-inTrain,]
# dim(training)
set.seed(32343)
modelFit <- train(type ~.,data=training, method="glm")Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred# modelFit
predictions <- predict(modelFit,newdata=testing)
# predictions
cm <- confusionMatrix(predictions,testing$type)
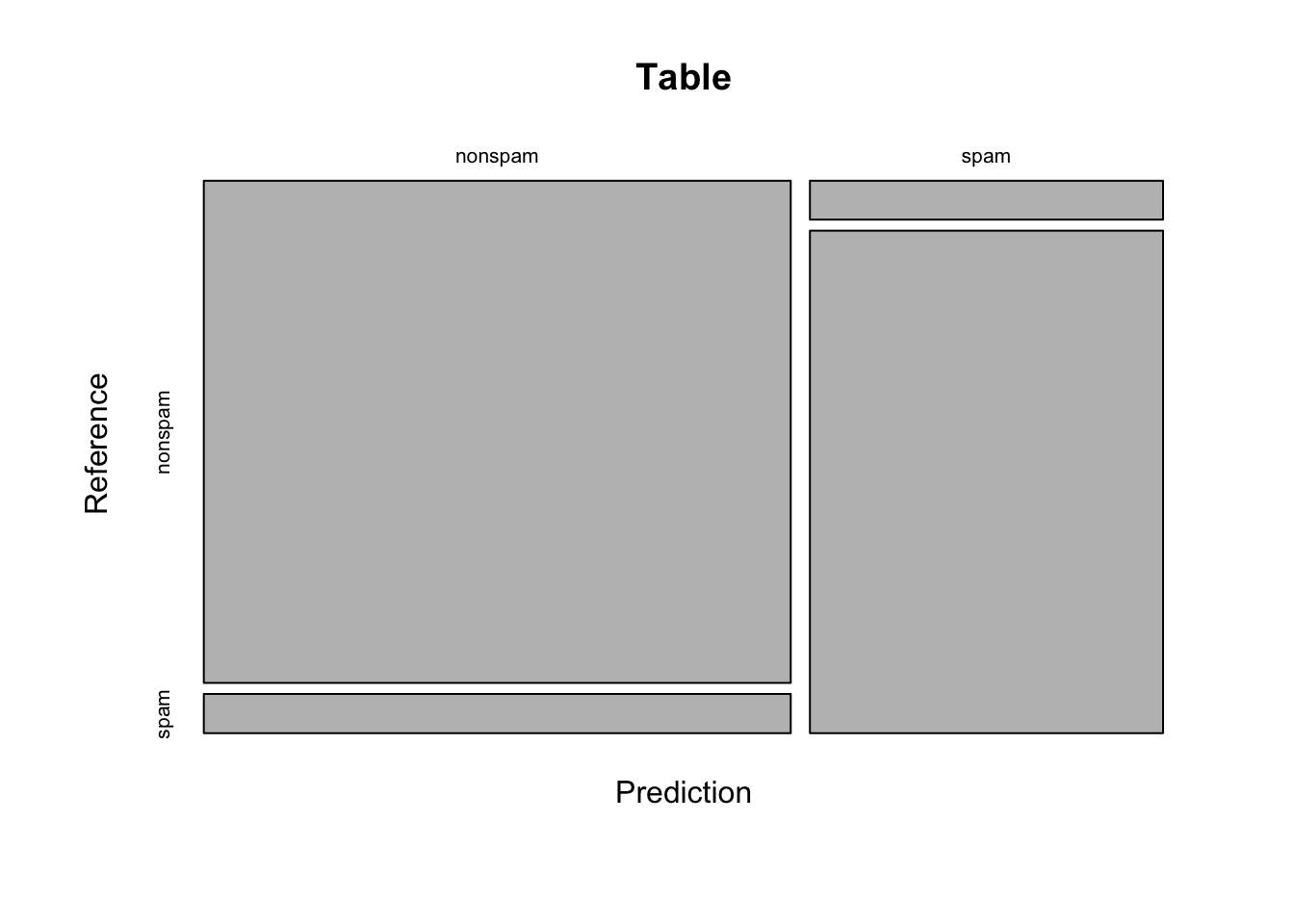
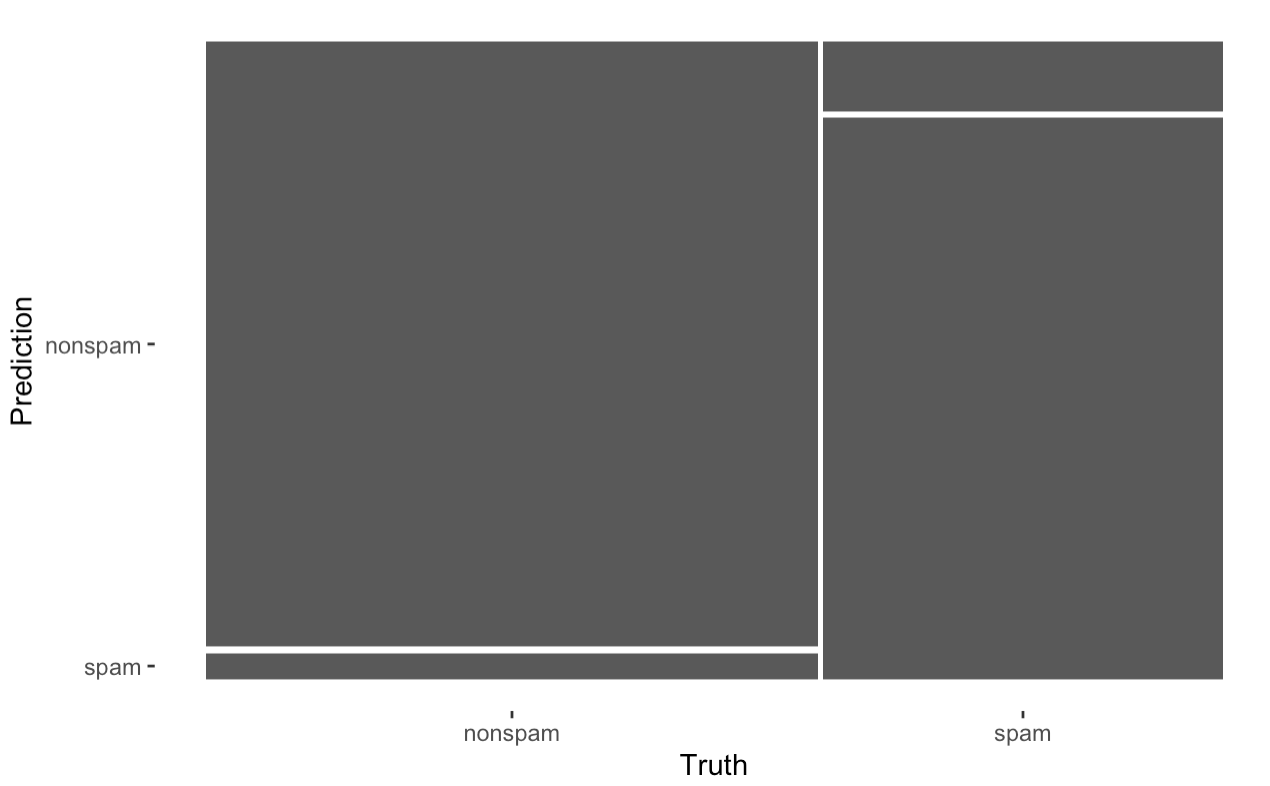
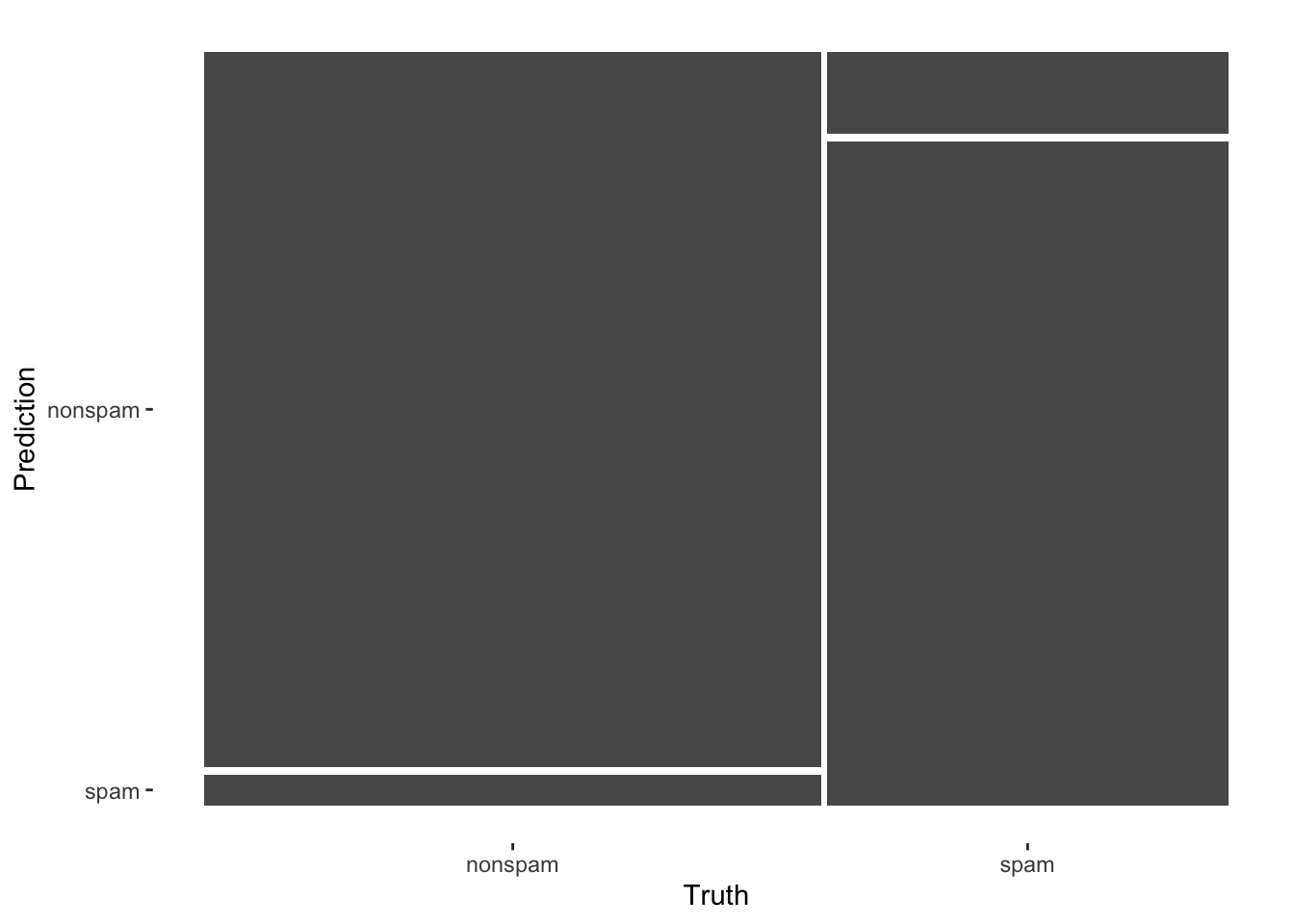
cmConfusion Matrix and Statistics
Reference
Prediction nonspam spam
nonspam 666 52
spam 31 401
Accuracy : 0.9278
95% CI : (0.9113, 0.9421)
No Information Rate : 0.6061
P-Value [Acc > NIR] : < 2e-16
Kappa : 0.8476
Mcnemar's Test P-Value : 0.02814
Sensitivity : 0.9555
Specificity : 0.8852
Pos Pred Value : 0.9276
Neg Pred Value : 0.9282
Prevalence : 0.6061
Detection Rate : 0.5791
Detection Prevalence : 0.6243
Balanced Accuracy : 0.9204
'Positive' Class : nonspam
plot(cm$table,main="Table")